Within this page, the GerBI-GMB Workgroup Image Data Analysis and Management provides an overview of selected (*) image analysis software packages. In addition, the advanced user might also find some deeper insights on the Bioimage Informatics Search Engine (BISE) web page which is supported by the Neubias Cost Action.
(*) We’d like to point out that this list represents only a small fraction of available software. The selection is based on criteria like:
- Usability (for basic users)
- Open Source software
- Software which is mainly used by the community (based on the results of the German BioImaging Image Analysis Survey)
- Software which is supported by a large (developer) community, thus providing sustainability
- Exptertise/Focus of the Image Analysis Workgroup members
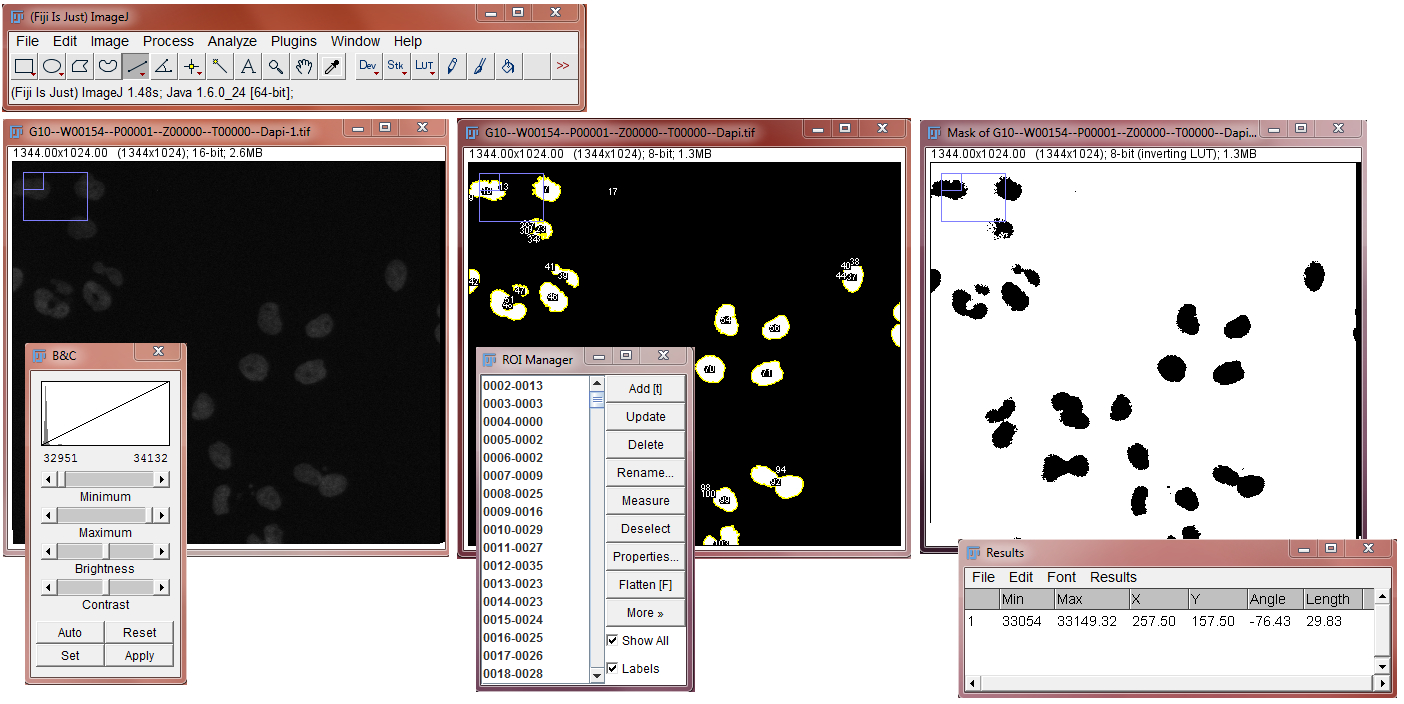
Category
- “Standard” Image Analysis
- Interactive representations
Example tasks
- counting nuclei & spots in 2D
- Object tracking in 2D (2D + t tracking)
- 3D object based co-localization
Appearance
Links
Fiji: http://fiji.sc/
Image J: https://imagej.net/
Category
- Object classification
- Image segmentation
- 2D/3D tracking
- Interactive representations
Example tasks
- Counting nuclei & spots in 2D
- Object tracking in 2D (2D + t tracking)
Appearance
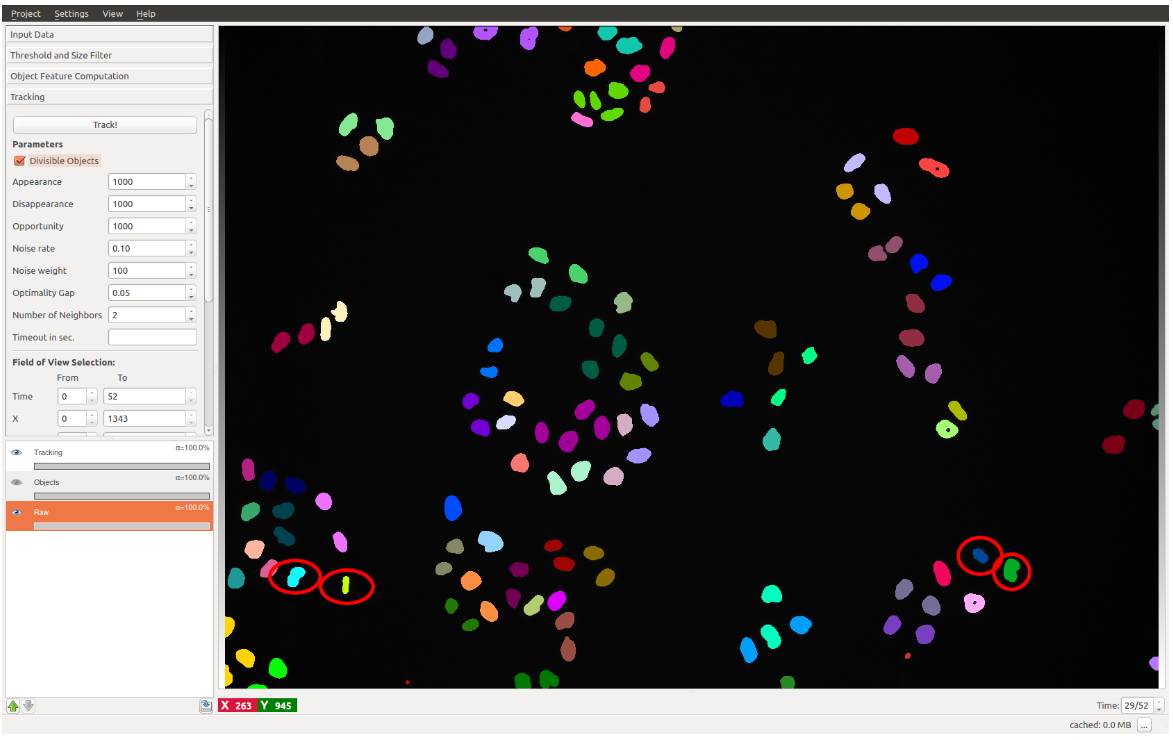
Link: https://www.ilastik.org/
Data pipelines
- “Standard” & High-throughput Image Analysis
- Interactive representations
- Statistics /Data exporation
Example tasks
- Counting nuclei in 2D
- Analyzing cytoplasm intensity in 2D
- Counting nuclei & spots in 2D
Appearance
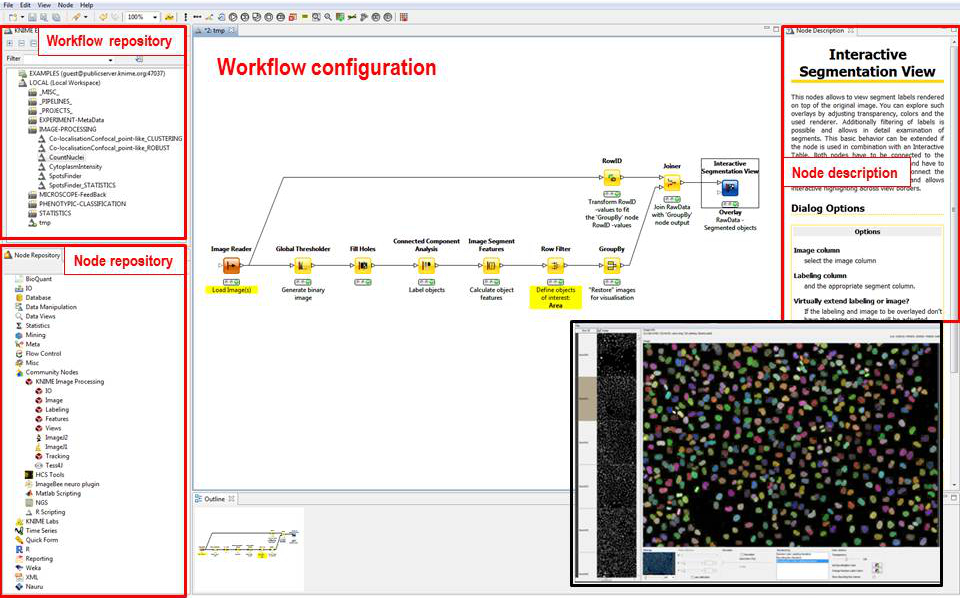
Link: https://www.knime.com/
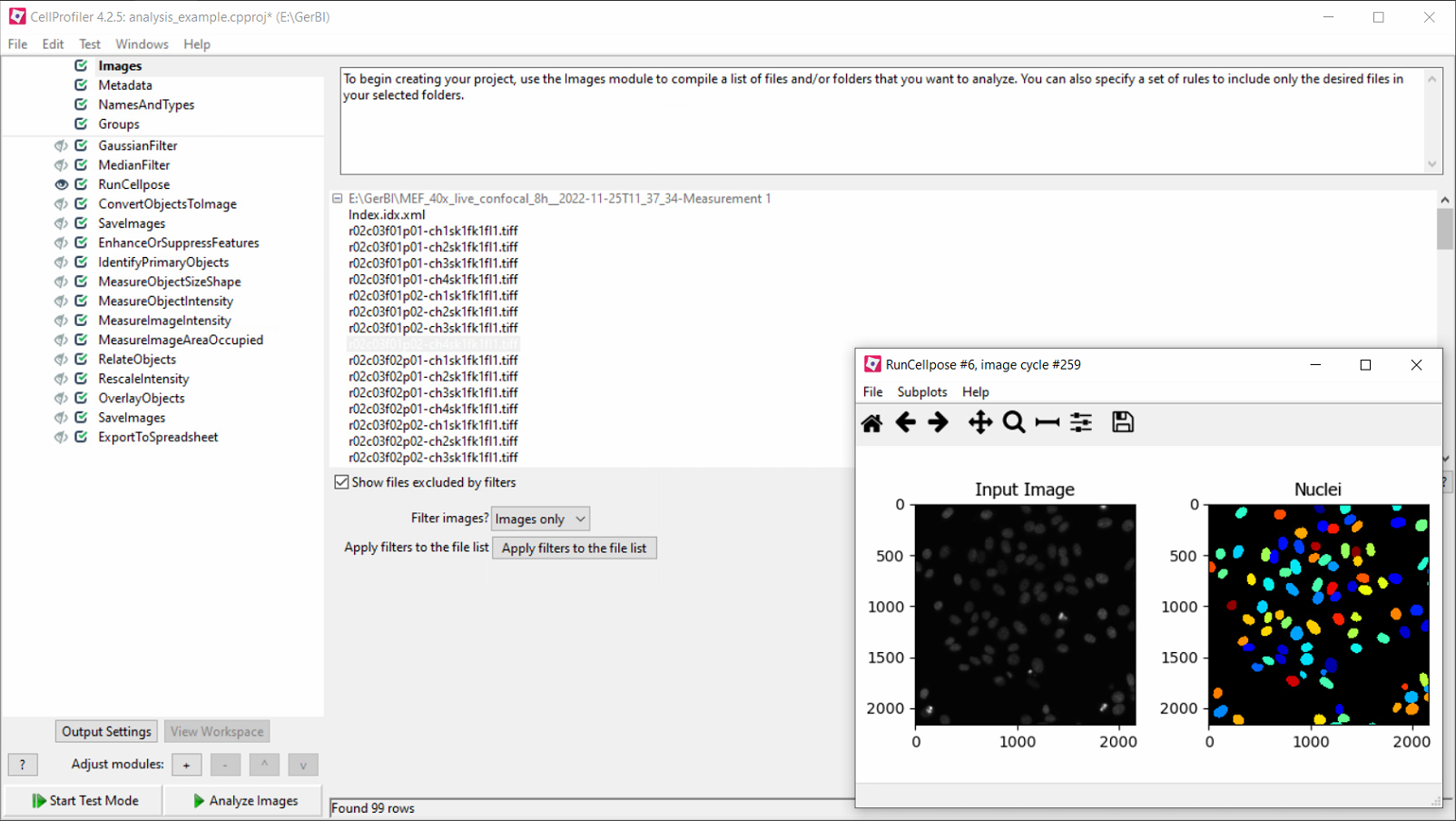
Category
- High-throughput image analysis
- High-content image analysis
- Statistics
- Interactive representations
Example tasks
- Segmenting and analyzing objects
- Cellular phenotyping
- Co-localization
- Object tracking
Appearance
Header image: Vater-Pacini corpuscles, masson goldner trichrome stain, finger berry human, Zeiss Axioplan Mot 2, Steffen Köhler, CAi, HHU

