Digital images in bioimaging for beginners
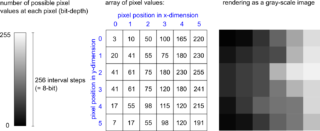
Back to: Microscopy File Formats Digital images for beginners How does the computer store a microscopy image? Photographs and microscopy images are recordings of real-world objects acquired with imaging devices. A microscopy image is thus a representation of the real object based on experimental setup and the device used (i.e., not "the" reality). The light that was detected during image acquisition is [...]
Basics of Imaging File Formats
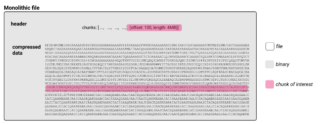
Back to: Microscopy File Formats Bioimaging File Formats explained The smallest piece of information underlying a computer is the distinction of two states (on or off), which can be represented as a binary digit (bit) with values 1 or 0. To store more complex information, more bits are combined. E.g., 8 bits in a row make up one byte (typically the smallest [...]
Bio-Formats – a translator of different file formats
Back to: Microscopy File Formats Translating Bioimaging File Formats with "Bio-Formats" The Open Microscopy Environment Consortium has created the so-called Bio-Formats library (Linkert et al., 2010, J Cell Biol) to enable the translation from a large number of proprietary file formats to the OME data model, generating OME-TIFF as a potential output file. Most files generated at the vendor-specific microscopes are written in proprietary, [...]
Next-generation file formats for bioimaging
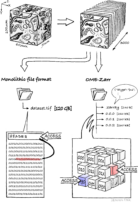
What are next-generation file formats (NGFF)? Structure of a classical file format As introduced in Bioimaging File Formats explained, microscopy image data can be written in various different file formats, mostly defined by the vendors of the microscopes used. Most of these files and also the "de facto" open community standard file "OME-TIFF" are classical file formats. I.e., these files are written as a series [...]