What are next-generation file formats (NGFF)?
Structure of a classical file format
As introduced in Bioimaging File Formats explained, microscopy image data can be written in various different file formats, mostly defined by the vendors of the microscopes used. Most of these files and also the “de facto” open community standard file “OME-TIFF” are classical file formats. I.e., these files are written as a series of bits in a row (binary data). The files in bioimaging include a header, i.e. the bits in the beginning which contain necessary information about the data stored in the file below in a specified way. This type of information in the header is called “metadata” and the way it is structured is the metadata format. The rest of the file contains the pixel data itself (or any other binary data if it is not an image). The pixel data is an array of numbers to store the values belonging to each pixel. This means that the information belonging to a specific spatial location within an image is somewhere in the big blob of binary data. So if we’re interested in only this part of the image, we are only interested in a chunk of the whole file. Accessing this chunk in a classical, i.e., “monolithic” file requires reading through the binary file until the chunk of interest is reached. In an example of a compressed file, this could be depicted like this:
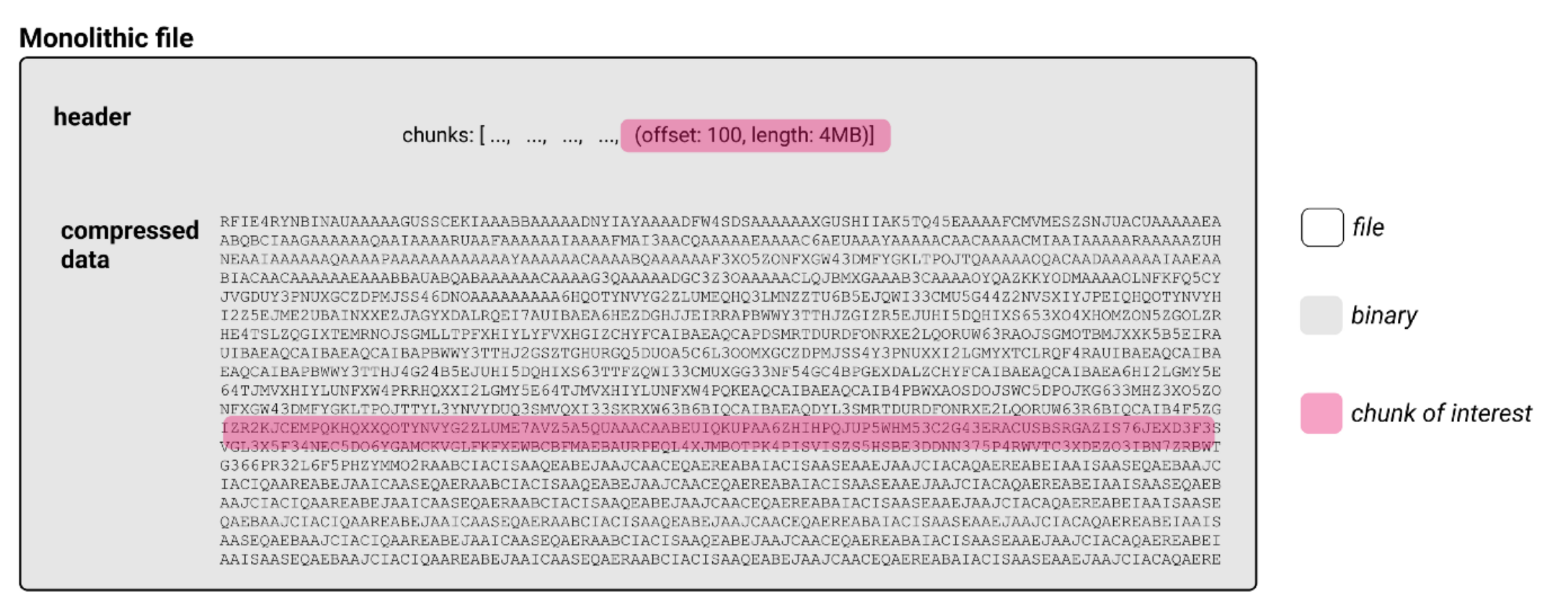
Simplified schematic view of a compressed monolithic file. Modified from an article by J. Moore.
What’s the issue with classical file formats?
To open and process image files, computers need to load the files into the computer’s memory. Large files can be for example subdivided into planes, which allows loading only the required planes of a stack. However, to load data inside a plane, the whole plane is loaded. Loading data across several planes and at user-defined angles would become more or less impossible since some files (e.g., in volume EM imaging, light-sheet microscopy, multiplexed imaging, etc.) can be really large (GB to TB size!). Processing of terabyte-sized image files is beyond the limits of what most scientists’ computers can do. Powerful workstations or high-performance computers offer solutions but they are neither widely accessible nor easy to use. Moreover, it is very difficult to access such files remotely when the data needs to be transferred from a location connected via a network. Furthermore, these classical files are not optimal for object-oriented storage like S3.
Why are next-generation file formats a solution?
An important difference between a classical file format and a next-generation file format (NGFF) is that the latter makes it easy to access and process only the part of an image that is of interest at a given time. This is achieved because the file structure is different. Instead of a so-called monolithic organization found in classical formats, NGFF allow to access chunks of the data in the file by breaking the file down to a multitude of small pieces that are logically bound together to represent the whole file. That means, a next-generation file format allows to store chunked N-dimensional arrays. Thus, a multi-dimensional image can be assessed along any dimension, loading only the chunks of interest. This feature also renders NGFF as “cloud-ready” formats, which allow streaming data chunks of a large file instead of the need to transfer the whole file. Find more information on NGFF provided by the OME team here or watch an talk by C. Tischer introducing NGFF for bioimaging here.
A graphical representation of the difference between monolithic, classical files and NGFF (here in the form of a Zarr directory) is depicted below in a comic by H. Falk:
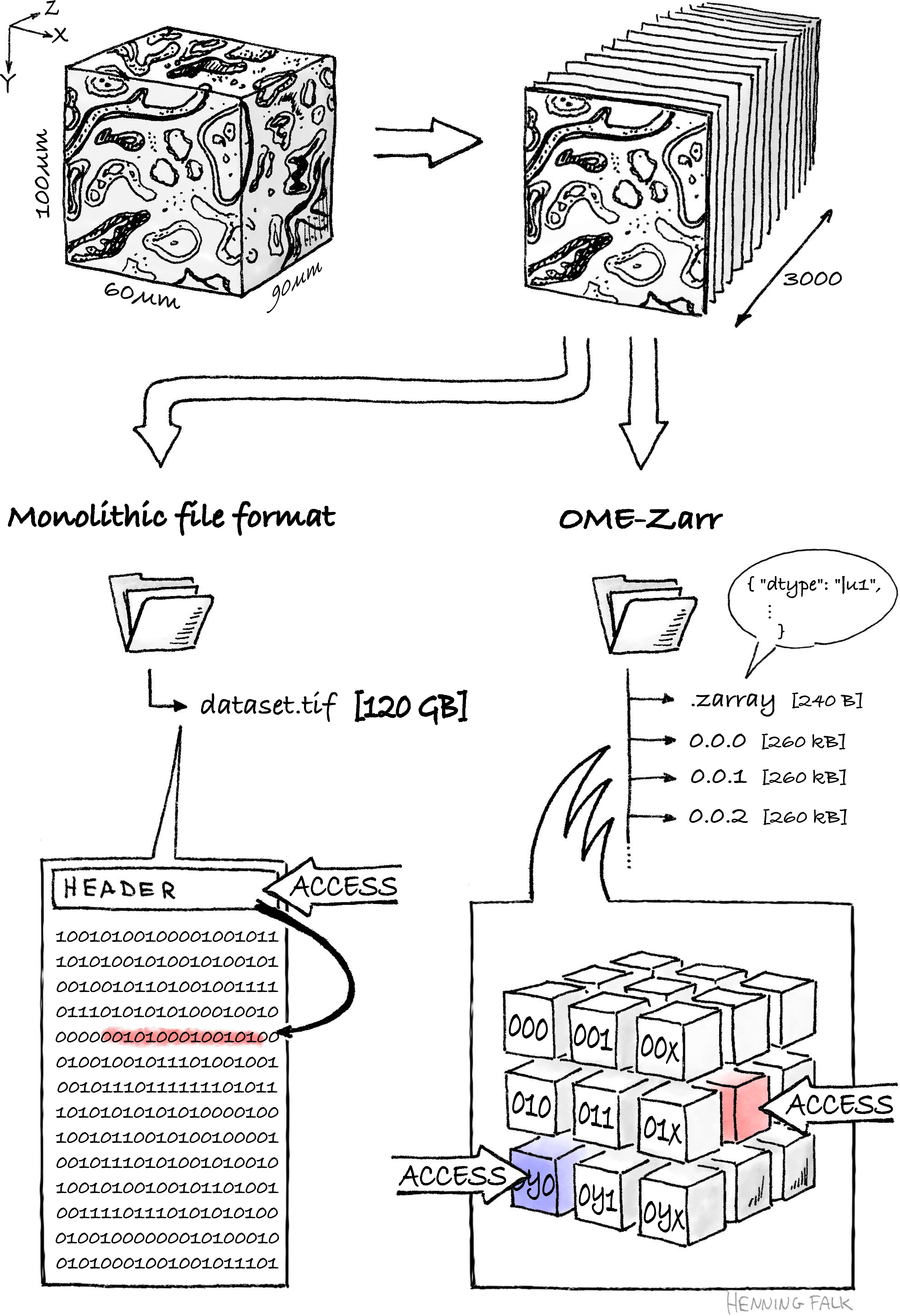
Falk, H. zarr-developers/zarr-illustrations-falk-2022 | Zenodo, 2022. URL https://doi.org/10.5281/zenodo.7037367 (accessed 21.11.22).
As depicted in the image above, data in a Zarr file format is structured into small, 3-dimensional chunks that can be directly accessed from the Zarr-directory. Hence, only the part of interest needs to be loaded instead of the full binary file. This feature allows NGFF data to be accessed in sufficiently small pieces so that streaming the data of interest over a network instead of transferring the whole file becomes possible.
Different implementations of a NGFF are:
- Zarr (forming the implementation of the OME-NGFF specification)
- n5 (which is used by the ImageJ community)
For the recent developments with respect to Zarr, please check out the recording of the Open Microscopy Community Meeting 2022 on Zarr and its visualisation and the 2023 publication on OME-Zarr by Moore et al.
On October 2nd, 2023, the development of OME-Zarr was in focus of a Technology Feature in Nature: How open source software could finally get the world’s microscopes speaking the same language (Nature, 2023)





