Back to: Microscopy File Formats
Digital images for beginners
How does the computer store a microscopy image?
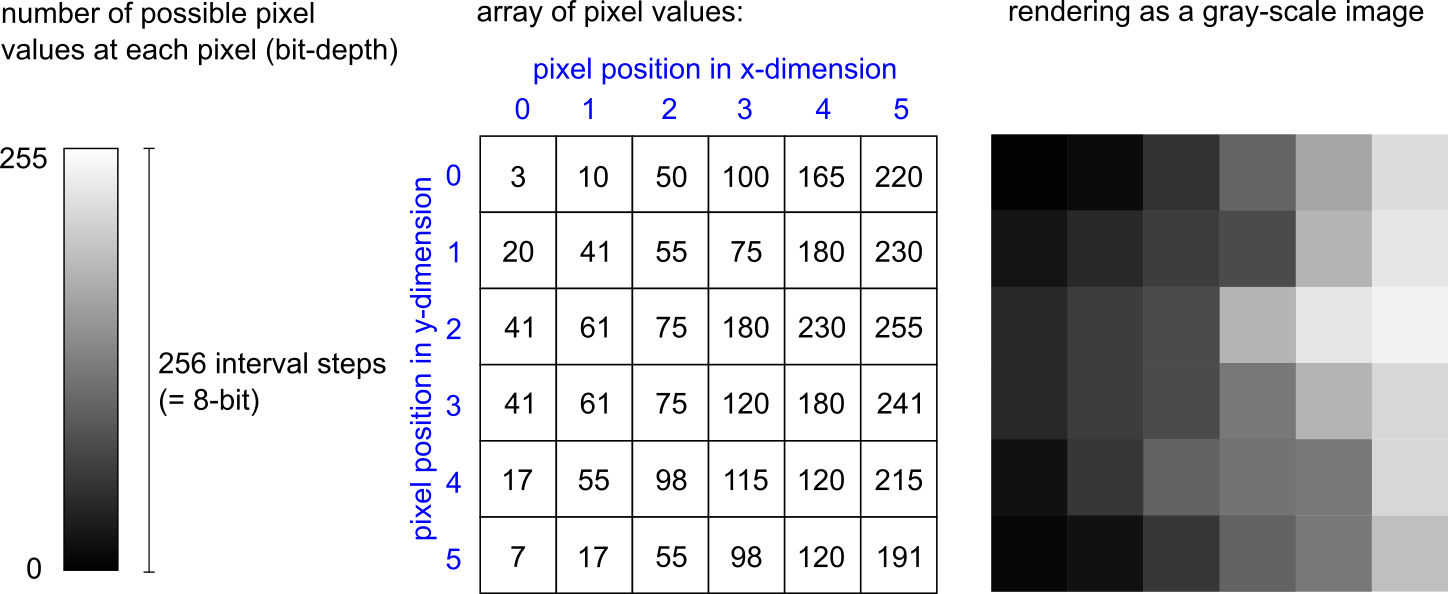
Photographs and microscopy images are recordings of real-world objects acquired with imaging devices. A microscopy image is thus a representation of the real object based on experimental setup and the device used (i.e., not “the” reality). The light that was detected during image acquisition is transformed into a digital representation of its intensity at a given spatial position within the recorded image. For this reason, microscopy images are raster images (as opposed to vector images) that store image information in the form of individual spatial points (pixels = “picture elements”). To represent such an image by a computer, the image file stores an array of numbers referring to the pixels. In the simple case, a 2-dimensional image contains a defined number of pixels that are arranged in two dimensions (x-dimension and y-dimension). Each pixel has a position (defined by x and y coordinates) and a value, which typically denotes the intensity of the light that was detected at that image position when the image was recorded. The value at each pixel is one within a range of possible pixel values (e.g., between 0 and 255), depending on the bit-depth. An 8-bit image stores one of 256 possible values at each pixel. A 16-bit image stores one of 65,536 possible values at each pixel, hence containing a higher quantity of intensity information that is more fine-grained. For displaying an image on the screen, the computer reads the position and the pixel value and renders the image by assigning a particular brightness to each pixel value. In a greyscale image, that means the range between “no brightness at all” (black pixel, mostly pixel value 0) and the brightest pixel (white, full brightness, mostly the highest pixel value) is divided into as many intvervals as the bit-depth defines. An 8-bit greyscale image is shown on the screen by assigning one of 256 possible brightness intensities to each pixel in the image, thus rendering it visually on the screen as shown in the figure below.
In addition to the pixel values and the positions, the image file contains data required to interpret or correctly render the image. For example, to make a distance measurement inside an image, it must be known what size of the real object that was imaged is contained in a single pixel (e.g., “pixel size = 300 nm”). These additional data about the data are called metadata, more specifically, technical metadata. The combination of pixel values and metadata is stored in an image file. The file format defines the specific way how these data are written in a file.
If you want to learn more about digital images and their representation and use in bioimage analysis, we recommend the NEUBIAS Image Analysis Training Resource, as well as the lecture on bioimage analysis by Robert Haase. Introductions to the basics of (classcial) imaging files are provided by Shaw & Hinchcliffe, 2013, and with a focus on medical imaging by Larobina & Morino, 2013. A comprehensive introduction to these principles is furthermore provided in textbooks, e.g., “Principles of Light Microscopy: From Basics to Advanced” (Springer, 2022).
The complexity of bioimaging data
Data acquired in bioimaging experiments can be quite complex. The reason is that the imaging modalities used can produce very different types of images, image sizes, image dimensions, etc. Consider an experiment in which cells are imaged in two dimensions (x and y), but consecutively over several time points (t), each time recording three different fluorescence channels (c). The image is a 4-dimensional array, that can become quite large depending on its dimensions and bit-depth. An example of particularly large image file are whole slide images, i.e., the digitization of a whole microscopy object slide with wide-field bright-light or even multi-channel fluorescence imaging. Such images typically reach sizes in the two-digit gigabyte range. The organization of the image in the file format has to be in accordance with technical capabilities of computer software to allow using it for the desired purpose. This includes organization in tiles and storage at multiple resolutions in the same file (pyramidal structure). You can find a non-technical explanation of these concepts here.
Special imaging modalities like Fluorescence Lifetime Imaging (FLIM) or Hyperspectral Imaging (HSI) record even more values at each pixel, increasing the complexity of the image. In many advanced imaging modalities, file sizes and complexity pose challenges to both storage space and processing of images (Ouyang, 2017).





