Back to: Image Data Repositories
Introduction to Image Data Repositories and Public Archives
Many bioimaging experiments are time-consuming and cost-intensive, and the acquired images are almost always subjected to image analysis to quantify image features of interest. Many imaging approaches thus produce large datasets, the value of which may extend well beyond the original research question that led to its acquisition. Therefore, sharing of original microscopy images can foster a more sustainable use of imaging data by means of reuse. The potential of public data sharing has been well demonstrated in other fields of research before. For example, the Protein Data Bank has helped to greatly increase our understanding of protein structure, function, and compound design for clinical use (Berman et al, 2000). The success of the PDB culminated in its contribution to the development of Alpha Fold in 2021 (Jumper et al, 2021; Jones & Thornton, 2022). The sharing of underlying data from Cryo-Electron Microscopy imaging in the Electron Microscopy Data Bank (EMDB) and the Elecron Microscopy Public Image Archive (EMPIAR) showcases where public image data sharing is readily possible today. Domain-specific imaging data repositories have been established where large amounts of specific data types accumulate (e.g., VirtualFlyBrain or XNAT Central). But there is a large heterogeneity of imaging modalities and data formats in light microscopy. Often, depositing image data to public repositories is not (yet) standard practice (Schmidt & Hanne, 2022). Sharing the original data underlying published findings from a research article (instead of declaring the data availability upon request) increases trust and facilitates that readers and reviewers are able to fully understand and – if desired – reproduce the results presented. Therefore, bioimage data management should include data publication from a scientific article via deposition in suitable public archives. The need for public image archives in (light) microscopy has been called for by Ellenberg et al, 2018. The article distinguishes two general types of public archives for bioimaging:
„Data archives are long-lasting data stores with the dual goals of (1) faithfully representing and efficiently storing experimental data and supporting metadata, thus preserving the scientific record, and (2) making these data easily searchable by and accessible to the scientific community. An archive serves as an authoritative public resource for data, but it does not aim to synthesize datasets or make value judgments beyond assuring adherence to standards and quality. Archives make it possible to connect different datasets on the basis of common standardized elements, such as genes, molecules, and publications. A typical example of a biological data archive is the European Nucleotide Archive, which stores nucleotide sequences. Data archives often have a single global scope, even if they are provided by a distributed organization worldwide.“
(Taken from: Ellenberg, J., Swedlow, J.R., Barlow, M. et al. A call for public archives for biological image data. Nat Methods 15, 849–854 (2018). https://doi.org/10.1038/s41592-018-0195-8, License: http://creativecommons.org/licenses/by/4.0/)
This type of databases, „referred to as added-value databases, are synthetic: they enrich and combine different datasets through well-designed analysis, expert curation, and, where possible, meta-analysis. They typically provide integrated information and biological knowledge for a community of users. They may also include advanced functionalities such as question-oriented searches and queries, cross-comparison of datasets, and advanced mining and visualization. Examples of added-value image databases exist already, such as the Image Data Resource (IDR; https://idr.openmicroscopy.org/), which integrates cell and tissue imaging studies on the basis of genetic or drug perturbations and phenotype.“
(Taken from: Ellenberg, J., Swedlow, J.R., Barlow, M. et al. A call for public archives for biological image data. Nat Methods 15, 849–854 (2018). https://doi.org/10.1038/s41592-018-0195-8, License: http://creativecommons.org/licenses/by/4.0/)
What makes “a good image data repository”?
There are numerous places to store data on the internet where it can be made accessible for the public. So, why is creating public cloud folder somewhere to provide the data not enough?Data sharing is not an end in itself. To be useful and to add value publicly shared data must comply with a set of principles that make the data Findable, Accessible, Interoperable, and Reusable (FAIR) (Wilkinson et al, 2016). Various criteria exist that intend to make sure data repositories are fit for the job. For example, publicly archived data must be persistent, uniquely identifiable and immutable. A link to a cloud folder could be erased, the data could be deleted and who would be able to find the link in the first place if not pointed to it in a paper or similar? To get started, make yourself familiar with how to find the repository that is right for you. Read more about quality criteria for repositories and how to identify a good repository here: Finding and choosing a repository.
For (light) microscopy imaging data, specifically two repositories have been (and are continued to be) developed in recent years:
- The BioImage Archive (BIA), an archive-type repository (see also: featured post about the BioImage Archive).
- The Image Data Resource (IDR), an added-value database (see also: featured post about the Image Data Resource).
I3D:bio’s comments on data submission to a public archive
These recommendations are based on the QUAREP-LiMi-developed Checklist for publishing images and image analysis, see Schmied et al., 2023, Nat Methods.
The purpose of having data stored in a public archive should be to make the data underlying the scientific publication FAIR – findable, accessible, reusable, and interoperable. The mere statement that „data is available upon request“ is not recommended. Depositing the image used for a specific figure in a publication (e.g., Fig. 4 A and B, see sketch) is a minimal requirement. It should be avoided that lossy compressed images without the original channel information are used. For example, an RGB TIF of a multi-channel fluorescent image does not enable data users to perform original image analysis workflows with the data. Rather than single figure images the original images that have led to the scientific conclusion shown in a given figure should be provided. In some cases, the decision on what is the useful raw data might be a matter of debate (see Schmied et al.). For image deposition in a public archive, open file formats and a minimum set of metadata annotation are recommended. What‘s more, since most images are subjected to (often several steps of) processing and analysis, the image analysis workflows from the raw data to the published images and the derived quantifications should be reported.
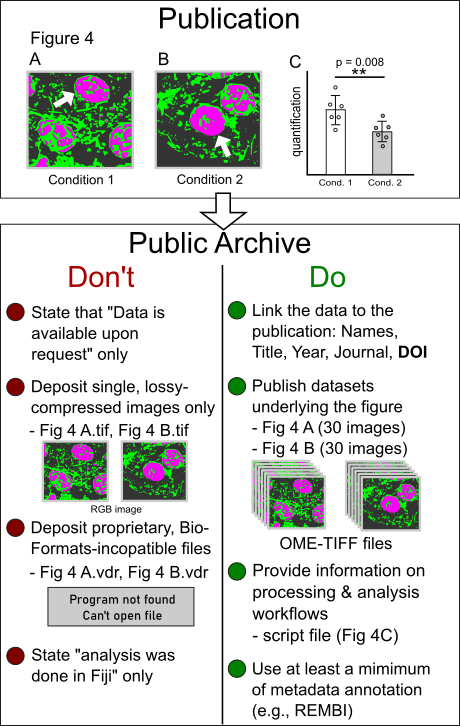
Simplified sketch exemplifying selected Do‘s and Don‘t for light microscopy image deposition in a public archive (for a comprehensive checklist, see Schmied et al., 2023, Nat Methods):





